|
>>
|
No. 27835
[Edit]
File
166892430793.png
- (443.30KB
, 4296x2492
, Structure Results 1048test3 1k_run_55 ( K = 5 ) ma.png
)
>>27833
>there was a confident implication that the "lack of evidence" is evidence against a connection.
Which is, ironically, the single greatest scientific fallacy of all time and which is the first you learn of. I think articles are being written very dishonestly these days, and tacitly approved by other groups by way of them simply not addressing the problem. These days it seems scientific newspapers just say whatever the fuck they want whole actual geneticists quietly publish completely contradictory evidence in the background.
Take this study by Rosenberg: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1310579/
The kind of work done in this study, along with all the work by Lazardis, is what every company like 23andMe, AncestryDNA, etc bases their tests on. We've been using computers to automatically sort and sift human genetic similarity to discover structure and relatedness for over two decades now. That's because the most important genes in determining human relatedness are autosomal DNA, which contain the genes for individual-specific phenotype, like height, skin color, hair color, eye color, general build, skull shape, whether you'll get multiple fingers or not, etc. Using a K clustering algorithm, where a computer is told to try and blindly sort samples of information into a number of groups K, we can find the population structure of human , animal, and plant populations. It's a little complicated but basically, people descended from the same population, like say a continent, will share a very large portion of their ancestors. >99% in a lot of cases. This means eventually the populations sort out and balance out around these ancestral makeups, and depending on how different two groups ancestors might have been, you can get a lot of drift. Sexual and natural selection also plays a large role.
Basically, if someone is descended from four europeans, three africans, and a native, it's not just going to look like some amorphous blob stuck somewhere between europeans, africans, and natives. It's going to be very clear for the most part which segments are from european ancestors and which are from africans or natives, usually 98-99% of segments can be classified in this manner. Also, people from certain populations that clearly have very relatively recent ancestors will be very autosomally similar. Any European, for example, will be incredibly similar to other europeans, and even the most distinct groups like sicilians or east russians are going to be within 90 - 95% similarity compared to other populations.
This is a simplification so don't just take it face value and think you understood it, but I've tried to be as honest and as objective as possible. Personally, I wouldn't use the word "race" because I don't see it as being the most up to date concept, but I also think that the ideas being described by race are pretty damn accurate and pretty close to the truth.
I won't get into major basal ancestries but it is simply known now that all human ancestry can be divided up into a handful of major archaic ancestries which correspond incredibly heavily to the variation discovered in humans by the early, "racist' human anthropologists. I mean, a race by any other name is still a race, right?
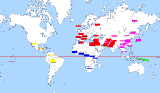
Here's the 5 main clusters you will identify in a 3D population clustering PCA, mapped out onto the locations of the countries they were taken from. Each box is a series of lines, each line showing the ancestry of one person, and the different colors in that line representing the proportions of the different clusters it assigned their genetics to. So you can see how some lines spike up a bit differently and that shows that individual have more or less of a specific ancestry than is common. It's a common format used for showing sampled and K-clustered genetic data, so that it can be easily visually sorted and distinguished in one graph.
|